Software
KVFinder
The KVFinder software is deprecated.
We published a more recent software, called parKVFinder.
parKVFinder
The identification and characterization of binding site is a major challenge in computational biology. Biomolecular interactions that modulate biological processes mainly occur in cavities throughout the surface of biomolecular structures. Computational tools to detect these cavities play an essential role in structure-based drug design. In the data science era, structural biology has benefited from the increasing availability of high-order biostructural data resulted from advances in structural determination and computational methods. In this scenario, we developed parKVFinder, a highly versatile and easy-to-use parallelized software for cavity detection and characterization. parKVFinder provides accurate, fast and efficient steered detection and spatial characterization of biomolecular cavities, with a set of intuitive and customizable parameters, which users can interact via a graphical user interface (GUI) or a command-line interface.
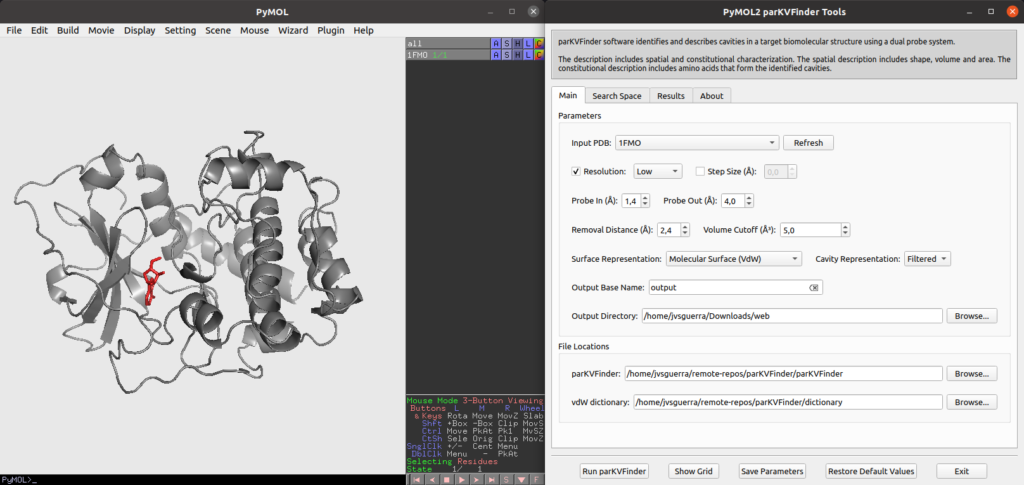
parKVFinder GUI overview
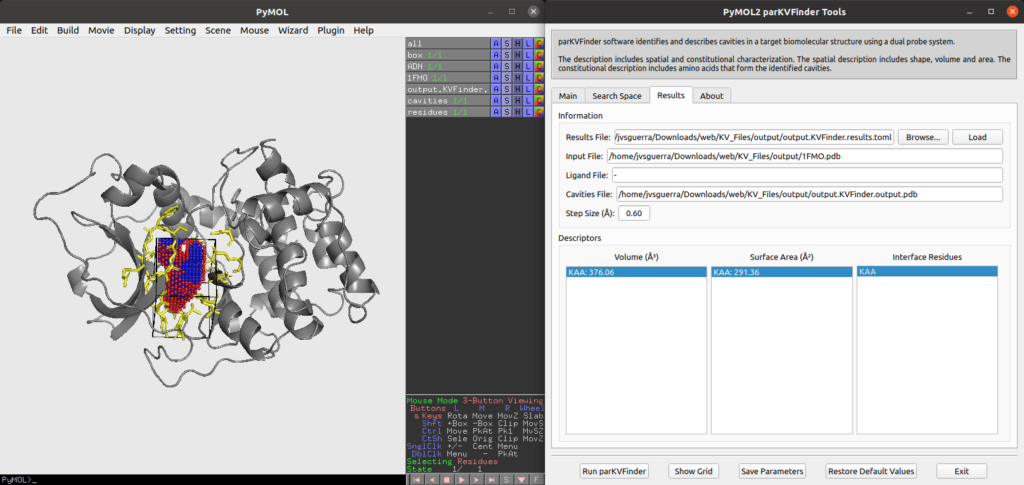
Example of parKVFinder results
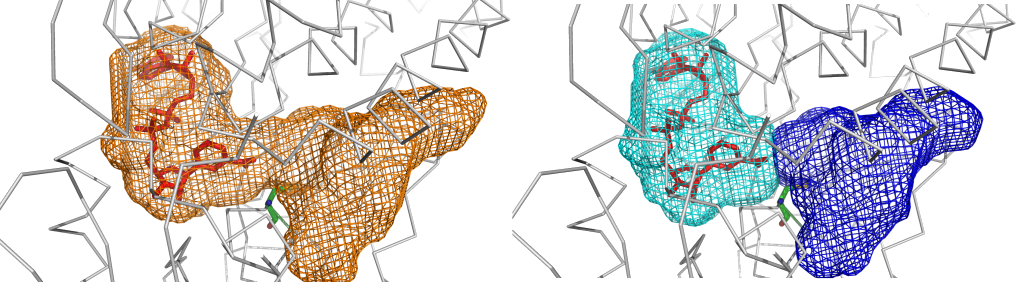
Space segmentation feature
ParKVFinder’s source code is freely available to the scientific community.
A Linux/macOS version is available on this GitHub repository, while a Windows version is here.
A complete documentation for parKVFinder software can be found here.
Please read and cite the original paper ParKVFinder: A thread-level parallel approach in biomolecular cavity detection (10.1016/j.softx.2020.100606).