Software
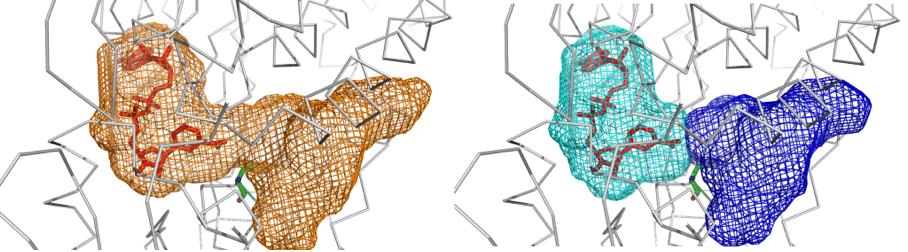
Cavity detection and characterization
The identification and characterization of binding site is a major challenge in computational biology. Biomolecular interactions that modulate biological processes mainly occur in cavities throughout the surface of biomolecular structures. Computational tools to detect these cavities play an essential role in structure-based drug design. Structural biology has benefited from the increasing availability of high-order biostructural data and large sets of atomic models, which require tools to balance accuracy and speed.
KVFinder
The KVFinder software, referred in a paper published in 2014, is deprecated.
We published more recent software, parKVFinder (2020) and pyKVFinder (2021).
parKVFinder
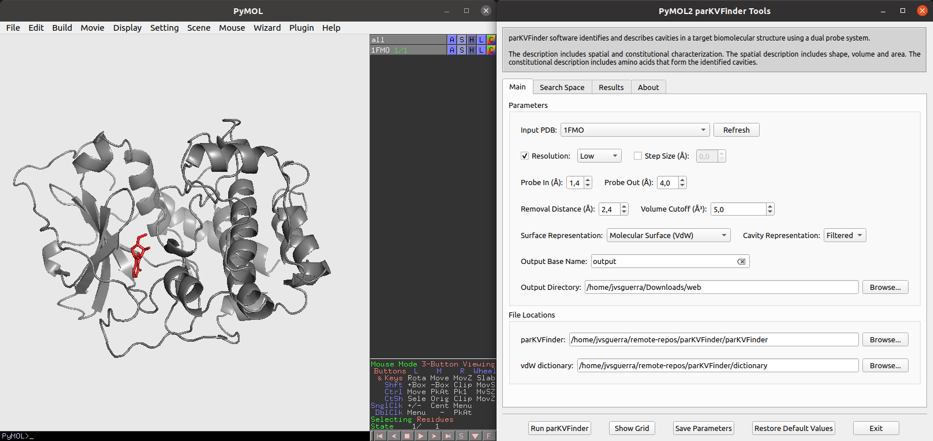
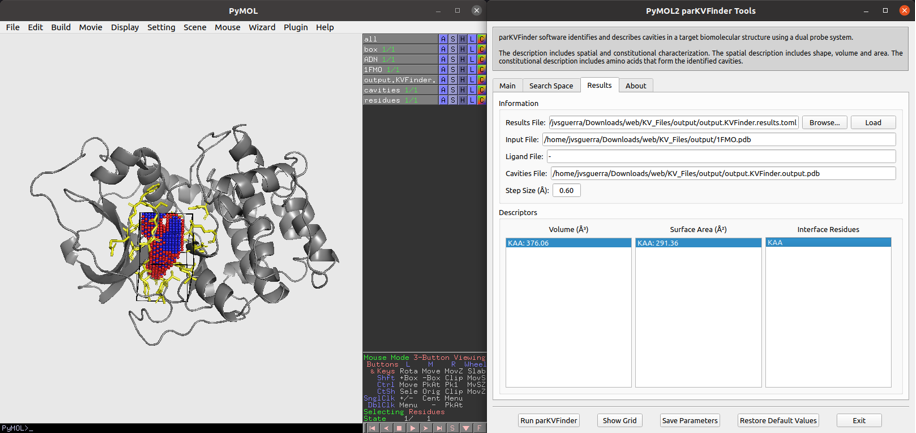
Parallel KVFinder, parKVFinder, is open-source (GPL v3.0) software designed for the detection and spatial characterization of any type of biomolecular cavity. ParKVFinder inserts the target biomolecule in a 3D grid divided by regular voxels and applies a dual-probe algorithm, originally implemented in KVFinder. parKVFinder provides accurate, fast and efficient steered detection and spatial characterization (shape, volume, area and surrounding residues), with a multithreaded parallelization implemented with OpenMP. Cavity detection relies on a set of intuitive customizable parameters, which user may interact via a graphical user interface (GUI) or a command-line interface.
A Linux/macOS version is available in this GitHub repository, https://github.com/LBC-LNBio/parKVFinder, while a Windows version is in this GitHub repository, https://github.com/LBC-LNBio/parKVFinder-win. Documentation and tutorials can be found at https://github.com/LBC-LNBio/parKVFinder/wiki.
Please read and cite the original paper ParKVFinder: A thread-level parallel approach in biomolecular cavity detection(10.1016/j.softx.2020.100606
pyKVFinder
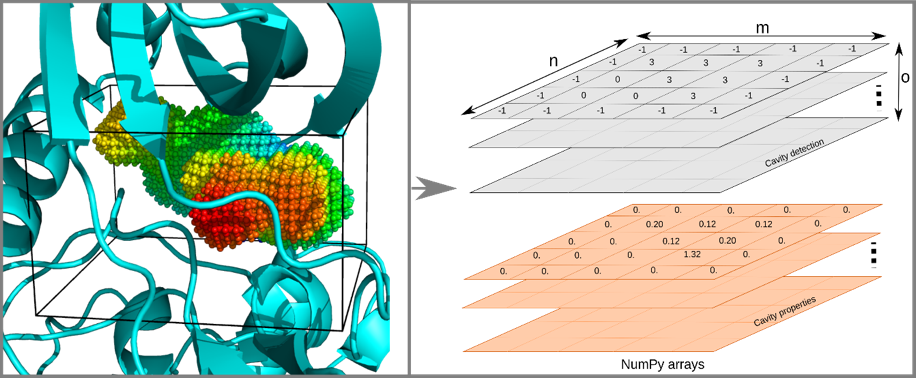
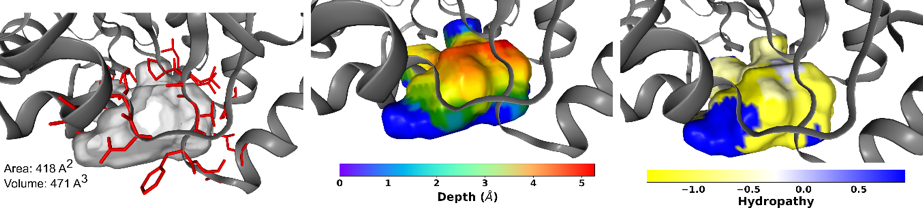
In the high throughput cavity analysis scenario, pipelines require efficient scripting routines built on easily manipulated data structures. To cover this requirement, we developed Python-C Parallel KVFinder, pyKVFinder, an open-source (GPL v3.0) Python package to detect and characterize cavities in biomolecular structures for data science and automated pipelines. In pyKVFinder, the target biomolecule is inserted into a regular 3D grid, which is stored as an N-dimensional array (ndarray). To detect cavities, pyKVFinder uses a dual-probe algorithm that scans the biomolecular structure, as described in KVFinder and parKVFinder.
Besides cavity properties such as volume, area and interface residues, which are stored as Python dictionaries, pyKVFinder computes cavity depth and hydropathy. Like cavity points, these spatial and physicochemical properties are stored as Python ndarrays and can be visualized using Python molecular visualization widgets such as NGL View. Thus, pyKVFinder facilitates biostructural data analysis with scripting routines in the Python ecosystem and can be building blocks for data science and drug design applications.
Hence, experienced users requiring scripting routines are encouraged to use pyKVFinder due to its improved performance, while newcomers should prioritize parKVFinder due to its simplicity of installation and execution.
pyKVFinder is available in this Python Package Index (PyPI) repository, https://pypi.org/project/pyKVFinder and this GitHub repository, https://github.com/LBC-LNBio/pyKVFinder. Documentation and tutorials are available at https://lbc-lnbio.github.io/pyKVFinder.
Please read and cite the original paper pyKVFinder: an efficient and integrable Python package for biomolecular cavity detection and characterization in data science (10.1186/s12859-021-04519-4).